Microscopy & Microanalysis 2022 in Portland (OR)/USA (July 31 - August 4, 2022)
Abstract
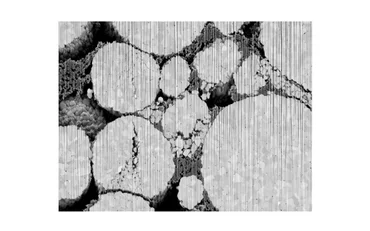
Performance, reliability, and safety of lithium-ion batteries play an important role in many industrial and consumer-oriented applications. The structure of a battery at the micro- and nano-scale greatly influences these properties and optimizing them must begin by understanding the structure at these scales. Detailed images of battery cathodes at the nano scale can be obtained through FIB-SEM devices. In these images, active material and binder are easily differentiated and even the composition of the grains can be investigated.
By their nature, FIB-SEM devices come with a disadvantage: the slice wise-obtained images show the current slice but also areas that are located behind pores, if the scanned sample is not infiltrated before imaging. Infiltrating samples is often avoided because it is a time-consuming task and might even change the structure of the material. For numerical simulations and geometrical analysis accurate 3D images of the sample are required. Therefore, the segmentation must be able to correctly differentiate the phases and to assign an area of a slice precisely to the current foreground or to the background. Adding to this challenge, the foreground is often affected by curtaining artifacts created by the Ion Beam.
Classical threshold or watershed-based segmentation methods often struggle with the task of segmenting FIB-SEM images correctly. In this study, these methods are compared on a FIB-SEM scan of a cathode provided by Zeiss with the more recent approach of trainable segmentations [1]. For the trainable segmentations, the performance of different machine learning-based approaches are compared. Specifically, we investigate a Boosted Tree [2] based method, a 2D U-Net [3], and a 3D U-Net [4] implemented in the GeoDict software. We show that the 3D U-Net performs best. The 2D U-Net creates good results when comparing only single slices but introduces discontinuity in the stacked direction of the slices. The training of the Boosted Tree method is much faster and requires less manually labeled data, but does not perform well in segmenting foreground and background of the FIB-SEM image.
[1] I. Arganda-Carreras, V. Kaynig, C. Rueden, K. W Eliceiri, J. Schindelin, A. Cardona, H Sebastian Seung: “Trainable Weka Segmentation: a machine learning tool for microscopy pixel classification”, Bioinformatics, Vol. 33, Issue 15: 2424–2426, 2017, https://doi.org/10.1093/bioinformatics/btx180
[2] T. Chen and C. Guestrin: XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD '16). Association for Computing Machinery, New York, NY, USA, 785–794, 2016, https://doi.org/10.1145/2939672.2939785
[3] O. Ronneberger, P. Fischer, T. Brox: “U-Net: Convolutional Networks for Biomedical Image Segmentation” Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, LNCS, Vol. 9351: 234--241, 2015, http://dx.doi.org/10.1007/978-3-319-24574-4_28
[4] Ö. Çiçek, A. Abdulkadir, S. S. Lienkamp, T. Brox, O. Ronneberger: “3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation” Medical Image Computing and Computer-Assisted Intervention (MICCAI), Springer, LNCS, Vol. 9901: 424-432, 2016, http://dx.doi.org/10.1007/978-3-319-46723-8_49